Celebrating Ten Years of Human Induced Pluripotent Stem Cells
Image courtesy of Tomo Saric, Medical Faculty, University of Cologne, Germany
2017 marks the 10th anniversary of the first human induced pluripotent stem cells (HiPSC) and the year of ECACC’s global launch of its collections of over 1,000 HiPSC cell lines (HipSci and EBiSC).
In 2006 Shinya Yamanaka’s Lab in Kyoto, Japan published landmark research that showed that stem cells with properties similar to embryonic stem cells (ESCs) could be generated from mouse fibroblasts by the introduction of four genes1. Takahashi and Yamanaka designated these cells “Induced pluripotent stem cells” (iPSC). Then a year later 2007, the lab reported that a similar approach could be used for human fibroblasts and that by introducing a few factors, human iPSCs (HiPSC) could be generated 2.
It soon became evident that HiPSC technology had two potential applications in medical science. Firstly in the field of regenerative medicine, with the dual ideas that a patient’s own cells might be re-programmed for autologous transplantation to correct disease states and also the generation of banks of tissue matched HiPSC could be generated as “off-the-shelf” treatments for many patients.
The second and perhaps more immediate application of HiPSC is in the field of understanding the mechanisms of diseases and drug discovery. HiPSC offer the potential of modelling diseases in tissue culture flasks, on the lab bench in a way never before imagined, simultaneously overcoming the ethical constraints posed by the use of human embryonic stem cells (HESC)3. These are the applications intended for ECACC’s iPSC Collection: HipSci
The first disease specific application of HiPSC was reported by Dimos in 20084, where HiPSC were generated from skin fibroblasts isolated from patients with amyotrophic lateral sclerosis (ALS) (also known as motor neurone disease (MND) or “Lou Gehrig’s disease”). The patients’ fibroblast cells were reprogrammed to HiPSC with retroviruses encoding KLF4, SOX2, OCT4, and c-MYC and subsequently differentiated to neuronal cells with a motor neurone phenotype and the patient’s genotype.
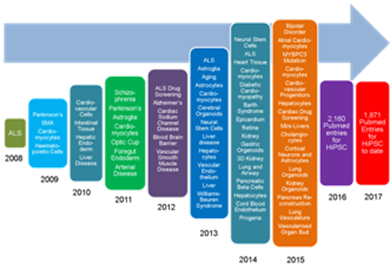
Since then the application of HiPSC has increased year on year at an increasing rate as shown in Figure 1. This figure uses data presented by Orlova and Mummery (2016)5 who recorded key developments in the application iPSC applications as reported in the literature. Since 2016 the number of citations of HiPSC in the literature is too numerous to list. This year’s running total is over 1,800 to date.
There is now a decade’s worth of literature demonstrating the application of HiPSC derived differentiated cell culture and organoid models for neuronal, cardiac, endodermal and mesodermal tissues. This literature well and truly demonstrates both the worth and potential of HiPSC as key reagents in the development of treatments and cures of many diseases where there are unmet clinical needs and for the establishment of physiologically relevant in vitro models for early toxicology testing in the drug discovery workflow. Cellular types that have been generated from HiPSC include: cardiomyocytes, vascular tissues, hepatocytes, optic cup and retina, pancreas, stomach and intestine, kidney, brain and airway epithelia amongst others. These models have allowed advances into the mechanisms and treatment of many diseases including ALS, Parkinson’s, liver metabolic disorders, arterial disease, spinal muscular atrophy (SMA), diabetes, Progeria, and Duchenne muscular dystrophy (DMD)5.
Figure 1
The key advances in disease modelling and iPSC differentiation are “snowballing” year on year at an increasing rate.(After Orlova and Mummery(2016)5)
Click here for larger version
With the wealth of HiPSC relevant literature being currently generated it is sometimes unclear to the reader on the effectiveness of a proposed iPSC derived model and whether it really does display physiological relevance. With that in mind, in the field of airway research, recognising the potential for HiPSC to generate disease specific models and to deliver personalised therapies Beers and Moodley (2017) set out a key set of criteria that would allow researchers to use as a checklist to determine if novel putative iPSC derived alveolar type 2 (ATII) cells are truly physiologically relevant. By setting out a clear set of molecular, morphometric, biochemical and functional metrics, Beers and Moodley have provided a useful gauge by which potential ATII models may be evaluated. Indeed, in their review the authors explore the advantages and shortcomings of many of the ATII models that have been proposed in recent years6.
There is no doubt that HiPSC cell lines have enormous potential to advance medical research even further in the next 10 years with even more promise offered by the combination of CRISPR CAS9 gene editing techniques7 however, if HiPSC based in vitro research is to be reproducible and valid, it is essential that these putative differentiated models are critically evaluated and compared to existing cell line derived systems and wherever possible, validated primary cultures.
References
- Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell126, 663–676 (2006).
- Takahashi, K. et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell131, 861–872 (2007).
- Yamanaka, S. Induced Pluripotent Stem Cells: Past, Present, and Future. Cell Stem Cell10, 678–684 (2012).
- Dimos, J. T. et al. Induced Pluripotent Stem Cells Generated from Patients with ALS Can Be Differentiated into Motor Neurons. Science321, 1218–1221 (2008).
- Orlova, V. & Mummery, C. SnapShot: Key Advances in hiPSC Disease Modeling. Cell Stem Cell18, 422 (2016).
- Beers, M. F. & Moodley, Y. When Is an Alveolar Type 2 Cell an Alveolar Type 2 Cell? A Conundrum for Lung Stem Cell Biology and Regenerative Medicine. Am. J. Respir. Cell Mol. Biol.57, 18–27 (2017).
- Hockemeyer, D. & Jaenisch, R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell18, 573–586 (2016).
