Diversity within the chicken gut
Chicken meat provides a popular low-carbon, healthier and more affordable alternative to meat from other livestock. As a result, globally this ubiquitous species now outnumbers the human population by three to one and accounts for a larger fraction of the planet’s biomass than all species of wild birds combined. The chicken gut microbiome underpins links between diet, health and productivity in poultry, while also acting as a crucial reservoir for zoonotic foodborne pathogens such as Campylobacter, Salmonella and E. coli, as well as for antimicrobial resistance genes. Despite its importance, the rich taxonomic diversity within this abundant habitat (which includes bacteria, viruses, archaea and microbial eukaryotes) remains largely undefined. To explore this key microbial community, we exploited two complementary approaches, which together delivered a survey of the chicken gut microbiome unprecedented in scale and scope.
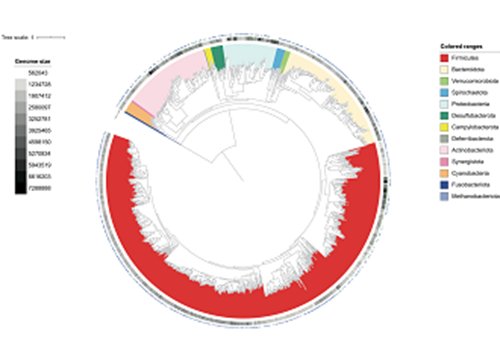
High-throughput culture of faeces from fifty UK chickens allowed us to create a catalogue of over 280 bacterial isolates, which were characterised by whole-genome sequencing. Subsequent phylogenetic analyses revealed that we had recovered thirty new species in pure culture, which we are pleased to deposit in the NCTC, as well as the DMSZ.
Our culture-independent investigations exploited shotgun metagenomics, drawing on our own samples plus an additional 580 publicly available chicken gut metagenomes from ten countries. Through sophisticated bioinformatics analyses, we created a catalogue of over 20 million microbial genes, recovered over 5500 metagenome-assembled genomes and documented over 800 bacterial species, nearly half of which were entirely novel.
Not content with simply documenting novel taxonomic diversity, we sought to create a stable and memorable nomenclature for all our newly discovered species. To achieve this, we created >600 well-formed Candidatus Latin binomials, which probably represents the largest assignment of taxonomic names to microbes in a single research project. In so doing, we have provided proof-of-principle for a scalable approach to the creation of Latin names that could be applied to the thousands of new species being recovered from other metagenomic projects.
Focusing on the genus Escherichia, we were surprised to discover remarkable diversity within this commonplace setting. In particular, we designated one of our cultured isolates as the type strain for a newly named species, Escherichia whittamii, so-called after the American microbiologist Tom Whittam, who pioneered our understanding of genetic diversity in E. coli and its relatives.
Our culture-based and metagenomic analyses have documented remarkable diversity in the bacterial, archaeal and bacteriophage components of the chicken gut microbiome. The isolates we have recovered alongside our genomic datasets will provide a key resource for future high-resolution taxonomic and functional studies on the chicken gut microbiome.
More information can be found on the Quadram Institute website: What lives inside the chicken gut?
Written by Rachel Gilroy
July 2021
