UK NEQAS for Microbiology: Molecular detection of SARS-CoV-2 EQA

The unprecedented pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causing Coronavirus disease 2019 (COVID-19) is posing challenges for SARS-CoV-2 testing laboratories in the UK and worldwide. These laboratories have to cope with increased testing demands to identify and maintain the required containment of affected individuals as well as delay and mitigation of spread of the disease. Molecular detection or antigen testing from respiratory specimens are the mainstay for the detection of SARS-CoV-2.
Other hurdles that some of these laboratories face are sourcing of reagents and testing kits and have resorted to run different assays on multiple testing platforms, adding another layer of complexity to the testing, quality control and validation of results, warranting a need to participate in an external quality assessment (EQA) program on a regular basis.
UK NEQAS for Microbiology has successfully introduced an EQA for molecular assays to provide an aspect of quality assurance to laboratories performing molecular detection of SARS-CoV-2, thus enabling them to monitor, evaluate and improve their performance. An EQA to monitor the performance of SARS- CoV-2 antigen detection assays is currently under development and likely to be available soon.
The UK NEQAS for Microbiology molecular detection of SARS-CoV-2 EQA was evaluated through two pilot studies and now runs on a monthly basis. The monthly distributions consist of two freeze-dried specimens and participants have to report on the presence or absence of SARS-CoV-2. The first pilot distribution was dispatched in May 2020 and the first live distribution on the 3rd August. There have already been three successful live distributions to date (29th Oct 20).
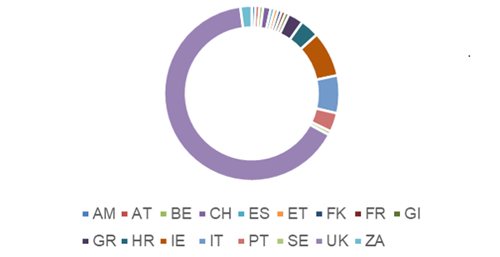
Interest in this scheme is increasing globally with participants from Armenia (AM), Austria (AT), Belgium (BE), Croatia (HR), Czech Republic (CZ), Ethiopia (ET), Falkland Islands (FK), France (FR), Gibraltar (GI), Greece (GR), Ireland (IE), Italy (IT), Portugal (PT), South Africa (ZA), Spain (ES), Sweden ( SE), Switzerland (CH) and the UK. The percentage of registrants vary between countries with 65% being from the UK as illustrated in Figure 1.
Participants registered for this EQA use varying methodologies that require different extraction, elution, reaction volumes as well as targeting different regions of SARS-CoV-2 genome (some assays having more than one viral gene target). The combinations of these factors greatly influence the sensitivity of the assays as shown in our reports.
From the data gathered, roughly, 49% of assays target the E gene, 22% target the ORF region including the RNA-dependent RNA polymerase (RDRP) gene, 22% the N gene and the remaining 7% the S gene. Figure 2 illustrates the genomic organisation of SARS-CoV-2, used as targets by the different assays.
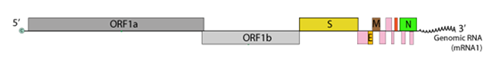
(Source: Swiss Institute of Bioinformatics https://viralzone.expasy.org/9076)
If your laboratory is running molecular assays for the detection of SARS-CoV-2, this EQA would certainly be beneficial to your laboratory. This EQA is supported by Department of Health & Social Care (DHSC), UK and NHS England.
Some of the benefits include a monthly specimen distribution with a two weeks period for examination and reporting results, followed by intended results the following day after a distribution has closed.
A detailed report follows within 10 days after the intended results have been published. This report highlights the performance of the different assays reported by the participants, at the same time being educational with up to date information on the evolution of COVID-19 globally.
More information about the UK NEQAS for Microbiology Molecular detection of SARS-CoV-2 EQA scheme and a recent report can be accessed through the following link: http://www.ukneqasmicro.org.uk/images/pdf/SARS-CoV-2.pdf
For further information and registration, please contact us by email: organiser@ukneqasmicro.org.uk
Written by Dr Sanjiv Rughooputh, UKNEQAS
November 2020
